APRANK: computational prioritization of antigenic proteins and peptides from complete pathogen proteomes
Abstract
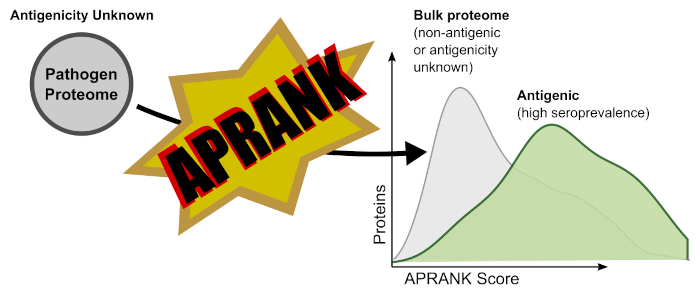
The long and boring (but academically correct) abstract is available at the publisher (read Abstract at Frontiers). Here we will provide a cooler version of what the paper is about. APRANK is an Antigenic Protein (and peptide) Ranker. To develop APRANK we used information on known and validated antigens and their peptidic epitopes (spoiler alert: this is all biased towards linear epitopes), and used it to train two models (one for proteins, to predict antigens; one for peptides to predict epitopes). APRANK takes a complete proteome as input, calculates a number of features for each protein and peptide, and outputs a score of how likely it is that the given protein or peptide is antigenic. APRANK has been trained with antigenic proteins and peptides from a phylogenetically wide diversity of human pathogens, and has been validated to works for bacteria, protozoa, and metazoan parasites (helminths).
APRANK is available to predict antigenicity but also for hacking and modifying.
We have made all code, data and trained models available. Code is under a simplified BSD 2-clause license, which is a permisive free software license. Feel free to reuse, retrain, and take APRANK to the next level.
If you read the paper, you will get pointers to all the relevant places, but in short:
- Code is at Github (https://github.com/trypanosomatics/aprank)
- this includes code to run APRANK on a new pathogen (using our pan-species, already trained models)
- as well as code needed to re-train APRANK
- Trained models are available at Dryad (https://doi.org/10.5061/dryad.zcrjdfnb1)
- due to size restrictions, these are made available separately here, and need to be placed in the corresponding directories in the APRANK code (once you check it out from GitHub)
- APRANK predictions for 15 human pathogens are in the Supplementary Materials (and also at Zenodo (https://doi.org/10.5281/zenodo.4939160)).
Hopefully everything is well documented in the code! Enjoy.